Appearance
Diffusivity of ellipsoid shell structure
For details about the effective diffusivity calculation, please see here.
Overview
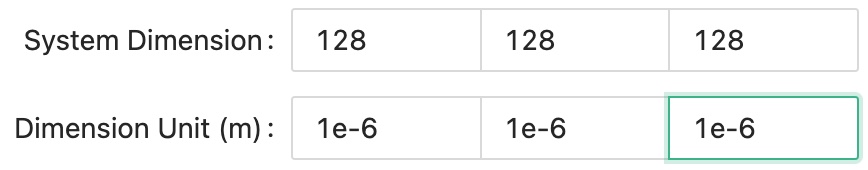
First, we will setup a cubic simulation with 128 microns along each dimension, that is 128 simulation grids times 1 micrometer for each grid.
Next, we will add three phases into the system, one for the matrix with lower diffusivity, and the other two for the ellipsoid shell and its inner part.
Lastly, we will setup a structure of ellipsoid shell.
Step 1: Fill Input
Follow the introduction to the input part in the user manual and make sure you fill all of the blank boxes in the following four sections.
Dimension
Output
Choose vti for the output format, since we may want to visualize with Paraview.
System
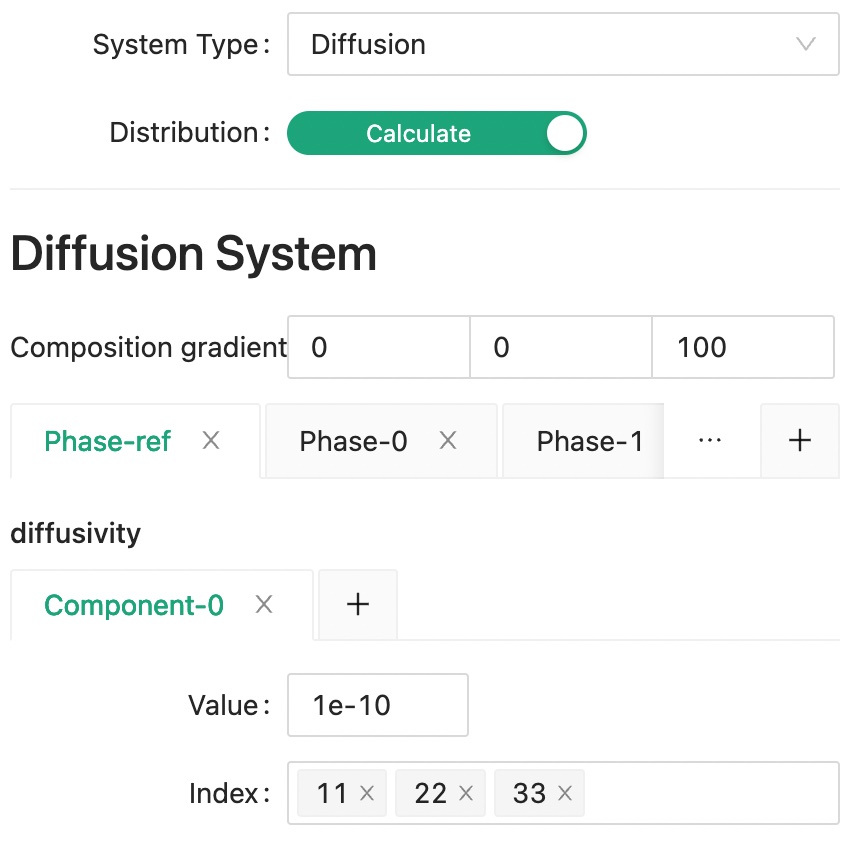
Choose "Diffusion" system for the System Type. Besides from the effective properties of diffusivity, we also want to calculate a field distribution on external applied concentration gradient, so we need to turn on the Distribution switch and set an Concentration gradient value.
Next, set the reference diffusivity value to be used in our solver.
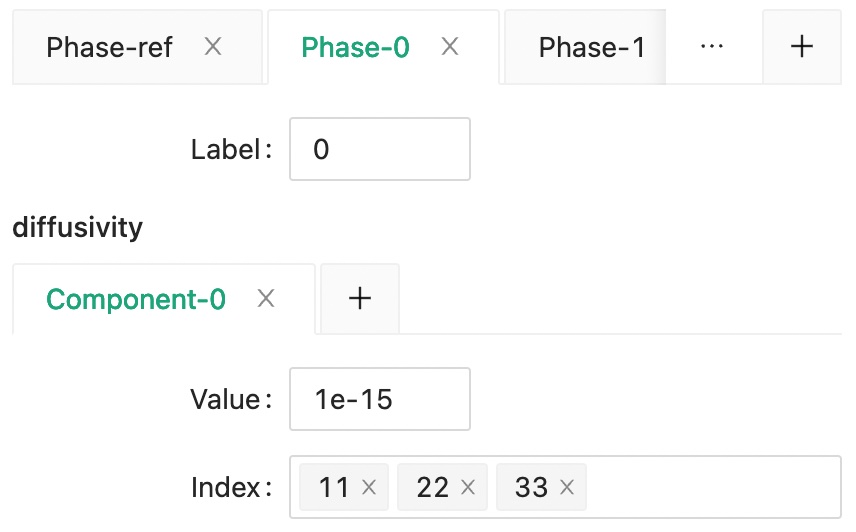
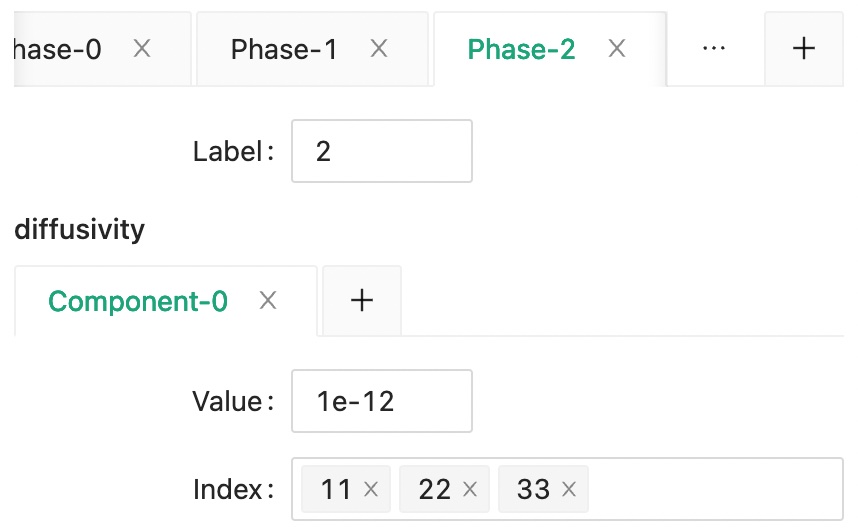
Then, set the diffusivity of matrix phase as phase 0.
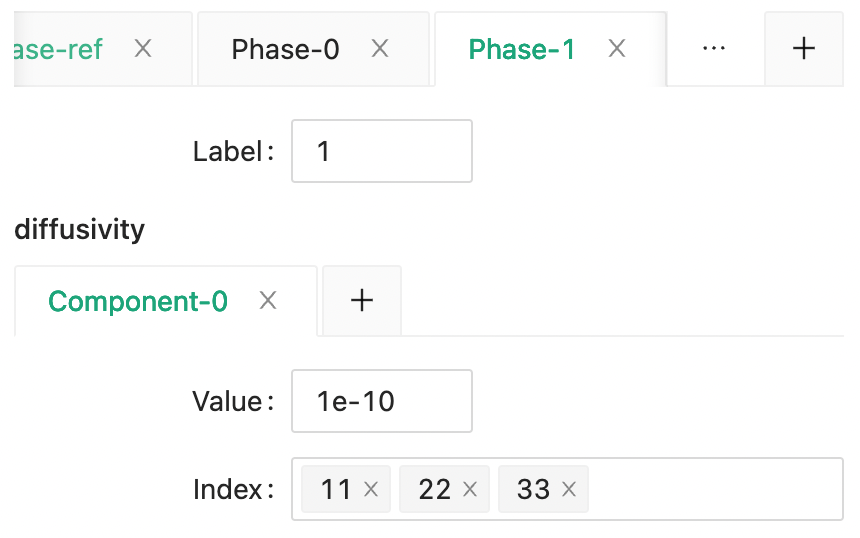
And the the diffusivity for phase 1 and phase 2.
Structure
Since we want to generate the structure within the effective properties simulation program, we should choose "Generate from xml file" for the Source type.
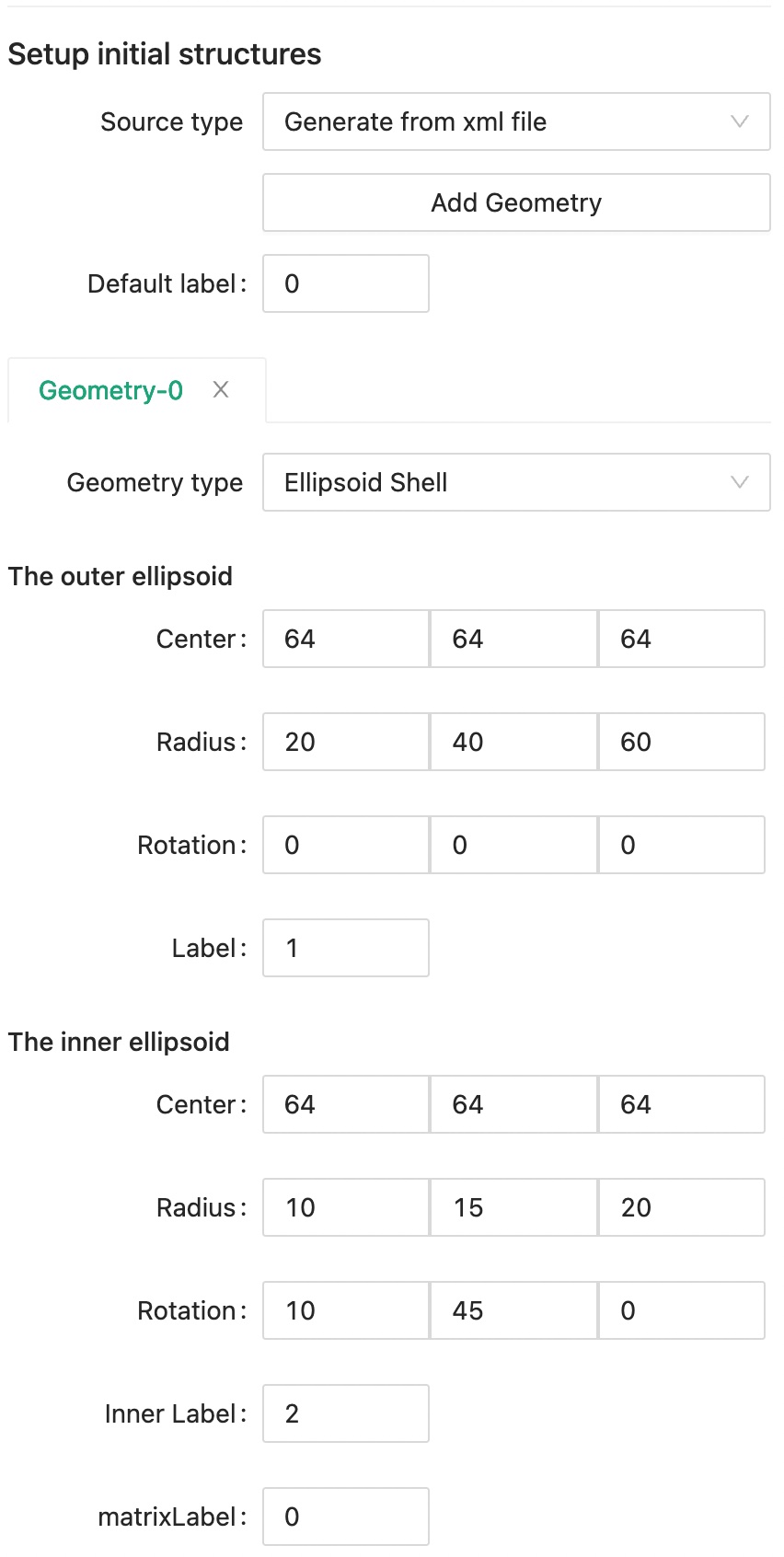
Click Add Geometry button to create a new geometry tab, and set label 0 as the Default label.
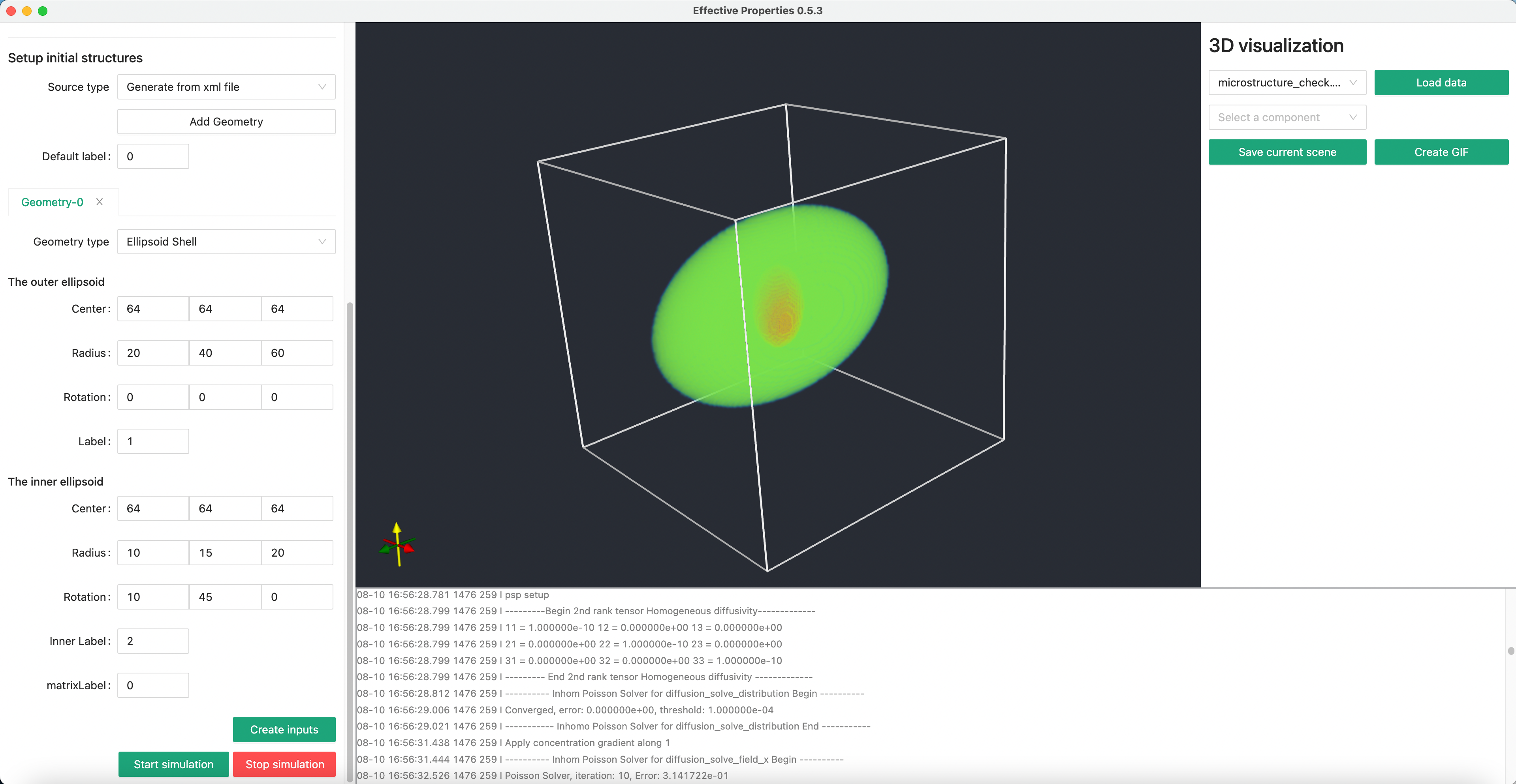
We want to create a ellipsoid shell at the center of the system.
We can set the inner and outer ellipsoid respectively as in the following figure.
Step 1.5: Or Import Input
If you don't want to enter all of the values manually, you may import the following input file directly into the software.
Save the following content into a local file, then click the File -> Import in the menu to read your saved file.
<input>
<dimension>
<nx>128</nx>
<ny>128</ny>
<nz>128</nz>
<dx>1e-6</dx>
<dy>1e-6</dy>
<dz>1e-6</dz>
</dimension>
<output>
<format>vti</format>
</output>
<system>
<type>diffusion</type>
<distribution>1</distribution>
<external>
<concentrationGradient>
<x>0</x>
<y>0</y>
<z>100</z>
</concentrationGradient>
</external>
<solver>
<ref>
<tensor>
<name>diffusivity</name>
<rank>2</rank>
<pointGroup>custom</pointGroup>
<component>
<value>1e-10</value>
<index>11</index>
<index>22</index>
<index>33</index>
</component>
</tensor>
</ref>
</solver>
<material>
<phase>
<label>0</label>
<tensor>
<name>diffusivity</name>
<rank>2</rank>
<pointGroup>custom</pointGroup>
<component>
<value>1e-15</value>
<index>11</index>
<index>22</index>
<index>33</index>
</component>
</tensor>
</phase>
<phase>
<label>1</label>
<tensor>
<name>diffusivity</name>
<rank>2</rank>
<pointGroup>custom</pointGroup>
<component>
<value>1e-10</value>
<index>11</index>
<index>22</index>
<index>33</index>
</component>
</tensor>
</phase>
<phase>
<label>2</label>
<tensor>
<name>diffusivity</name>
<rank>2</rank>
<pointGroup>custom</pointGroup>
<component>
<value>1e-12</value>
<index>11</index>
<index>22</index>
<index>33</index>
</component>
</tensor>
</phase>
</material>
</system>
<structure>
<matrixLabel>0</matrixLabel>
<sourceType>xml</sourceType>
<geometry>
<type>ellipsoid_shell</type>
<centerXOuter>64</centerXOuter>
<centerXInner>64</centerXInner>
<centerYOuter>64</centerYOuter>
<centerYInner>64</centerYInner>
<centerZOuter>64</centerZOuter>
<centerZInner>64</centerZInner>
<radiusXOuter>20</radiusXOuter>
<radiusXInner>10</radiusXInner>
<radiusYOuter>40</radiusYOuter>
<radiusYInner>15</radiusYInner>
<radiusZOuter>60</radiusZOuter>
<radiusZInner>20</radiusZInner>
<rotationXOuter>0</rotationXOuter>
<rotationXInner>10</rotationXInner>
<rotationYOuter>0</rotationYOuter>
<rotationYInner>45</rotationYInner>
<rotationZOuter>0</rotationZOuter>
<rotationZInner>0</rotationZInner>
<label>1</label>
<labelInner>2</labelInner>
<matrixLabel>0</matrixLabel>
</geometry>
</structure>
</input>
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
Step 2: Export Input
Click the Create Inputs button. Select the folder that you want to run the simulation in, and create a file named input.xml.
This step is simple but very important because the input.xml file is a prerequisite for starting any calculation.
Step 3: Run calculation
Make sure you have finished Step 2, you have to click the Create Input button, because it will both export the input.xml file and set the current working directory to that folder.
Then, you can click the Start simulation button to start the calculation. If you have not click the Create Input button and create the input.xml file, nothing will happen because the program does not know where to look for the input.xml file.
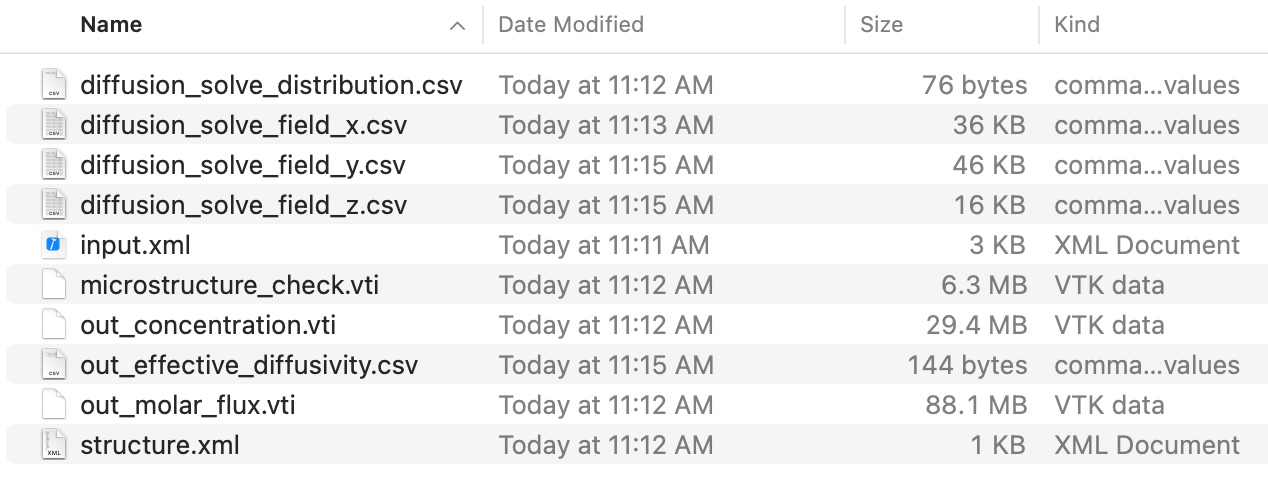
Step 4: Check Output
You will see the following output files in your simulation folder. Meaning for each of the files are explained in the Diffusion System.
Overall, there are two types of output data, vti files for 3D data, and csv files for tabular data.
Step 4.1: Check 3D data
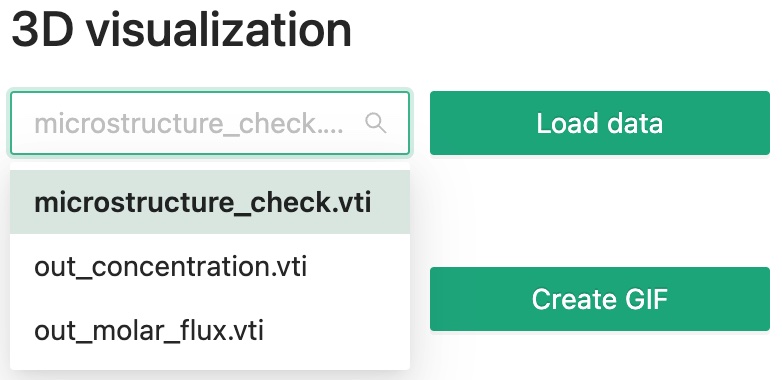
Within our software, you can quickly check a 3D vti data file.
Select the file you want to visualize using the dropdown menu, then click Load data button.
Then you will see something like this.
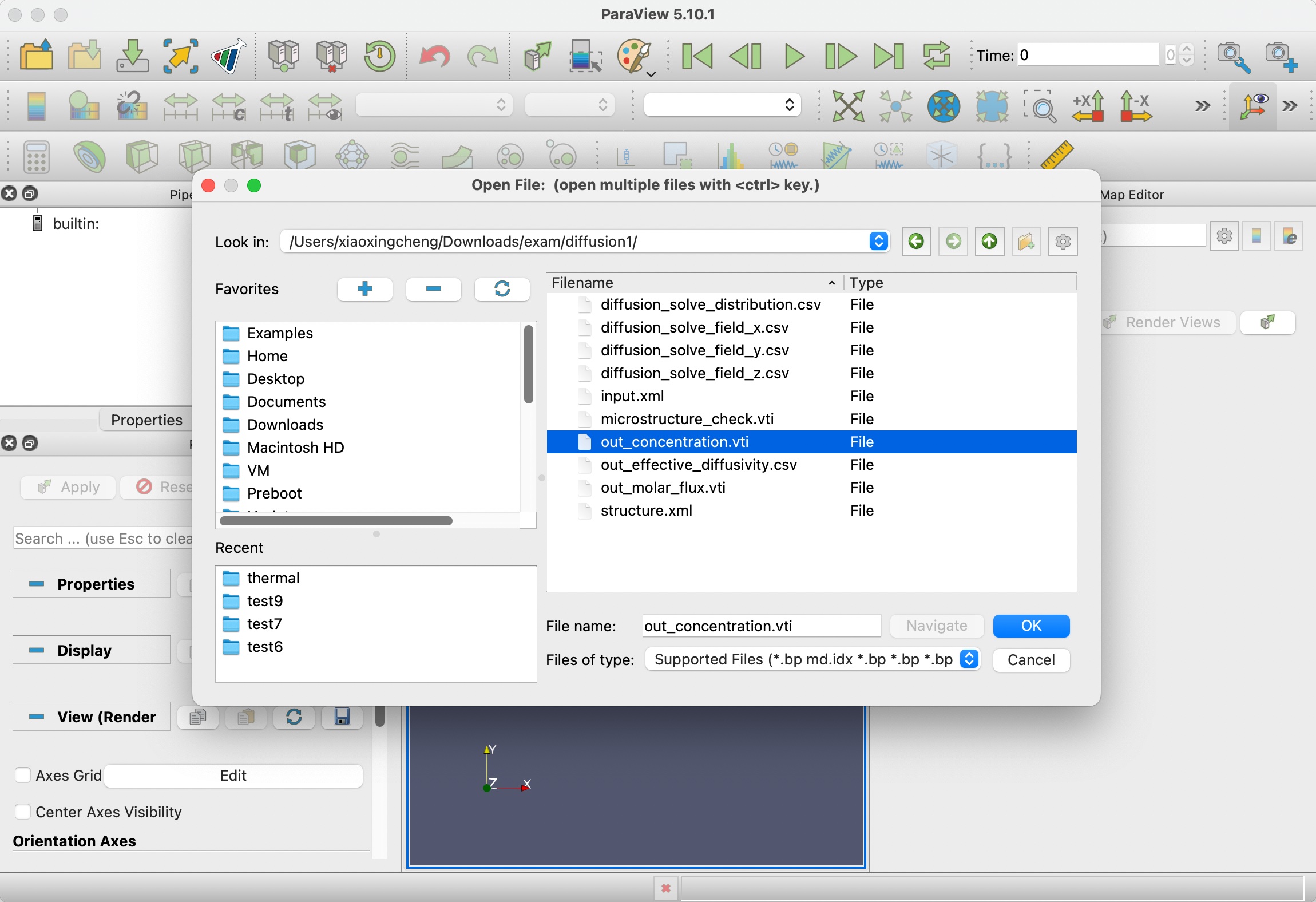
Step 4.2: Paraview
Next, we can try visualizing other files with Paraview. Click the first Open icon in the tool bar.
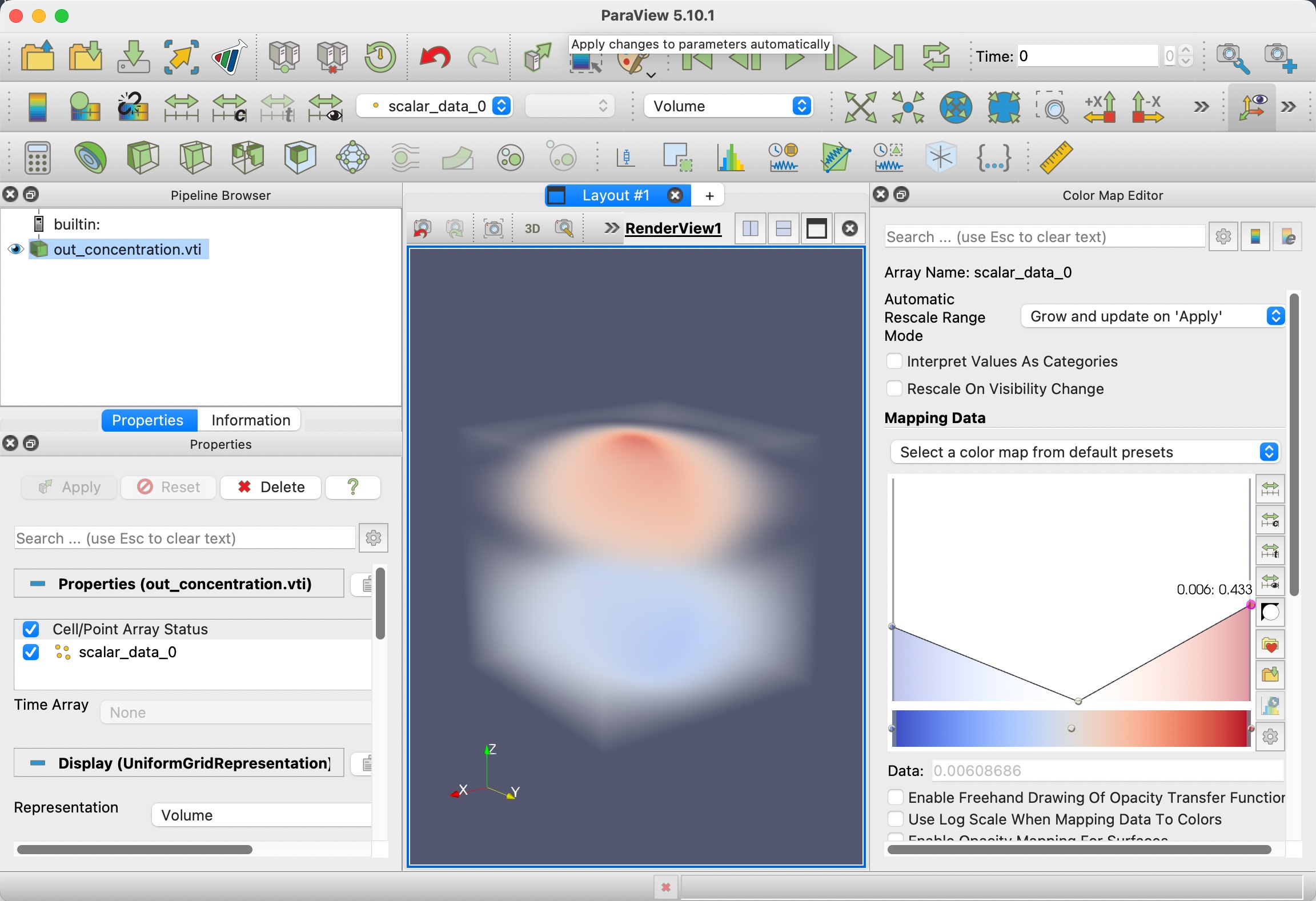
After the data is loaded, click Apply, then switch to Volume rendering and choose scalar_data_2, which is electric field along z for visualization.
We also need to tune the color lookup table to add some transparency to the data.
Step 4.3: Check tabular data
Though 3D data looks cool, the more important thing probably is still the effective permittivity, out_effective_diffusivity.csv.
| Index | 1 | 2 | 3 |
|---|---|---|---|
| 1 | +1.408238e-15 | +0.000000e+00 | +0.000000e+00 |
| 2 | +0.000000e+00 | +1.810113e-15 | +0.000000e+00 |
| 3 | +0.000000e+00 | +0.000000e+00 | +3.751247e-15 |